My early research studies centered on the development of an RNA sequencing method (Donis-Keller et al., 1977) and applications of the method (Rommelaere et al., 1979) when I was a graduate student at Harvard University in the laboratory of Wally Gilbert (who received a Nobel Prize in Medicine in 1979 for his work on developing a DNA sequencing method). While a research scientist at Collaborative Research, Inc., my research group focused on the construction of the first genetic linkage map of the human genome (Donis-Keller et al., 1987) and the application of genetic mapping methods to heritable disorders including cystic fibrosis (Knowlton et al., 1985), and neurofibromatosis type 1 (Stephens et al., 1989). During this time we also worked on developing DNA markers and using them to track the progress of bone marrow transplants (Knowlton et al., 1986) and using DNA sequences in general to distinguished human individuals from each other.

The Human Genome Project
Further development of the human genetic linkage map continued in my research laboratory at Washington University in St. Louis as part of the Human Genome Project sponsored by the National Institutes of Health (NIH/CEPH Collaborators Mapping Group, 1992).
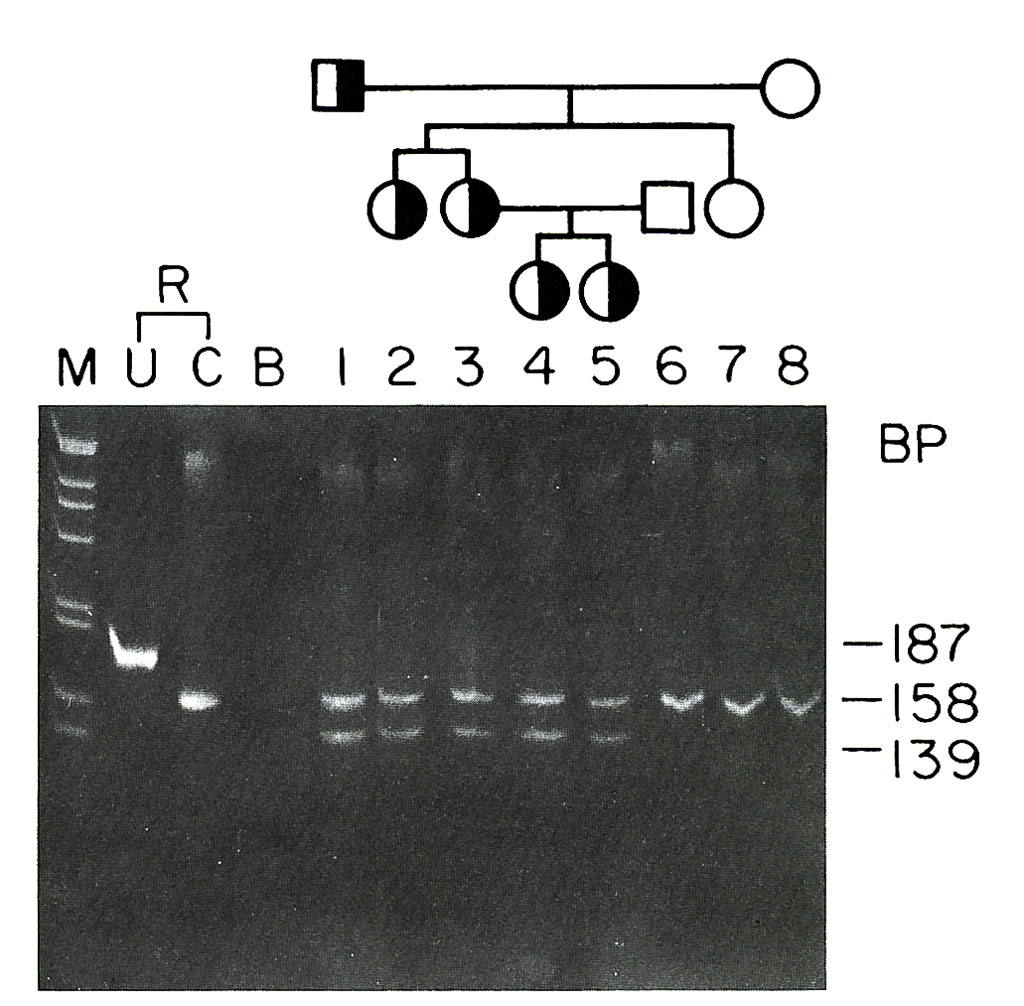
New applications of genetic mapping technologies to a variety of disorders including several heritable and sporadic cancers were also initiated. Research on the genetic locus of a form of thyroid cancer, Multiple Endocrine Neoplasia type 2A, led to the identification of the Ret proto-oncogene as the causative agent (Donis-Keller et al., 1993) and to predictive diagnostic tests for the disorder (Wells et al., 1994).
Thyroid Studies
Studies in my laboratory determined that another form of thyroid cancer, Multiple Endocrine Neoplasia type 2B was also caused by the Ret proto-oncogene (Carlson et al., 1994). Research in the laboratory on DNA markers for human telomeres, the ends of linear chromosomes, led to the identification and characterization of a large number of telomere-specific DNA markers (Vocero-Akbani et al., 1996) that found numerous clinical and basic research applications (Lengauer et al. 1993). My research group also participated in establishing and maintaining a breast cancer tissue resource for use in research and the development of treatments for the disease (Glass et al., 2001).
Moving to Olin
Upon moving to Olin College in Needham, Massachusetts, I became interested in studying the viruses of soil bacteria (mycobacteriophages). The overall goal of the research is to learn more about the evolution and diversity of the genomes of mycobacteriophages. Bacteriophages are the most abundant organisms in the biosphere and mycobacteriophages are an ideal resource for undergraduate research since they are readily available from environmental samples and they are not pathogenic to humans.
In the laboratory students isolated new phages and characterized them in terms of their morphology, growth characteristics, and their sequence organization.
In collaboration with the laboratory of Graham Hatfull and the Pittsburgh University Bacteriophage Genome Center, DNA sequences of the genomes of two Olin phages were determined, putative gene sequences identified and then compared to the panel of phages sequenced in the Hatfull laboratory (Hatfull et al., 2010). Students in my biology courses routinely isolate and characterize new bacteriophages as part of course-related laboratory experiences.
Recent work is focused on fungi biology and species conservation.